When I started my research lab at the University of Oregon in 2002, my first undergraduate student introduced me to Myotonic Dystrophy (DM). This student (Jeremy Logue*) was extremely motivated to study DM, he and his family were directly affected by the disease. Jeremy asked to study DM in my lab and initially I struggled with this idea because I worried that the slow progress of science would be frustrating for him and our inability to immediately help his family would be disappointing. His persistence won out and after reading the DM literature I realized my lab’s scientific expertise could make a difference in the field.
In 2002, the DM field had recently discovered that a toxic RNA (CUG repeats) was a major part of the disease mechanism. It was important to gain a better understanding of this toxic RNA. Part of my educational training included an approach (X-ray crystallography) that scientists use to determine the structures of biological molecules at atomic resolution. Structures of biological molecules have provided fundamental insights into how our bodies work at the molecular level. Importantly, crystal structures have also been used in developing drugs for many diseases. One such example is a crystal structure of a HIV protein that was used to guide the design of what is now a FDA approved HIV drug.
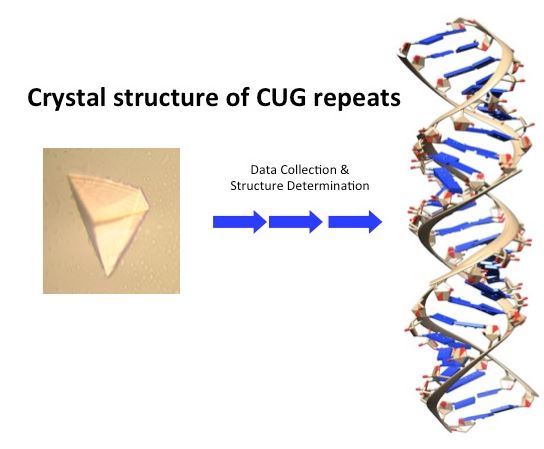
I realized we could potentially make a significant impact in the DM field by solving the crystal structure of CUG repeats. Jeremy’s project was to obtain crystals of purified CUG repeats that could be used to solve the structure. In one of those moments that make science so exciting, I remember Jeremy showing me a beautiful crystal of the CUG repeats (associated figure). In collaboration with Dr. Blaine Mooers* we solved the crystal structure of the CUG repeats and published our results in 2005 in the Proceedings of the National Academy Journal (http://www.pnas.org/content/102/46/16626.full). This structure has helped the field better understand the toxic RNA of DM and has been used to develop small molecules that are being tested as lead compounds for the development of therapeutics for DM.
Obtaining the crystal structure of the CUG repeats created significant enthusiasm in the lab and several PhD students and undergraduate students began working on DM projects. The results of these projects has lead to a better understanding of how the CUG repeats cause toxicity by soaking up proteins that should be regulating cellular processes. At my first DM meeting in 2007, I interacted with several people with DM and family members and these interactions had a profound impact on me. I went back to Oregon with the goal of pushing forward on our efforts to identify small molecule drugs (antibiotics) already known to interact with RNA that could potentially be repurposed for use by DM patients to reduce the toxicity of the CUG RNA. We published our first studies on these efforts in 2009, again in the Proceedings of the National Academy Journal (http://www.pnas.org/content/106/44/18551.full). These studies were a collaborative effort with my students Bryan Warf* (graduate student), Catherine Matthys* (undergraduate student) and Dr. Masayuki Nakamori and Professor Charles Thornton at the University of Rochester.
I was fortunate to work with excellent colleagues (faculty, senior scientists, postdoctoral fellows, graduate and undergraduate students) at the University of Oregon who all made significant contributions to our DM research efforts over the last 15 years. DM research at Oregon continues through members of the lab who chose to stay at Oregon for professional and personal reasons. I interact frequently with these colleagues and I am excited that they continue to study DM and are helping to move the field forward.
Current and future DM research directions
In 2015, part of my research group and I moved to the University of Florida to continue our DM research efforts. This move was motivated by the opportunity to team up with leading researchers and clinicians at the University of Florida’s College of Medicine in the Center for NeuroGenetics (http://neurogenetics.med.ufl.edu), with the goal of increasing our capabilities to make innovative discoveries in DM and related neuromuscular diseases (ALS and ataxias) and move these discoveries into the clinic.
Although we haven’t yet discovered a magic bullet for DM patients, the studies with drugs used for other diseases have revealed what we believe is a chink in the armor of DM. Our studies indicate that cells struggle to make the toxic CUG RNA. An analogy would be that making the CUG repeats is similar to a train climbing a steep mountain grade. Taking advantage of this weakness in DM, we are studying small molecules that block or hinder the ability of the cell to make the toxic RNA (or block the train from making the steep mountain climb), but importantly allow the cell to make other RNAs (trains not climbing mountains will continue on their travels). We are studying how these molecules inhibit the production of the toxic RNA and are developing and testing new molecules to identify more active molecules. Complementing these studies are two other projects in the lab. The first is we have developed an approach in collaboration with the Wang lab (http://neurogenetics.med.ufl.edu/faculty/dr-eric-wang/) to specifically screen for small molecules that exploit the chink in the DM armor. In the second approach we are designing synthetic proteins that we predict will block the production of the toxic RNA.
As we think about developing drugs to help those with DM, it is important to precisely measure how a potential drug is improving outcomes (i.e. muscle function) for the affected individuals. This is accomplished by measuring muscle function and changes at the cellular level in patients. The cellular changes are referred to as biomarkers and these can be used in both pre-clinical (i.e. cell models) and clinical trials to assess the activity of potential drugs. To help develop robust biomarkers we teamed up with Eric Wang and Charles Thornton at Rochester to sift through millions of sequences from DM patients to identify robust biomarkers that could be used for clinical trials. Mining this data and modeling the data with a cell system allowed us to identify biomarkers that can be broadly used in clinical trials and help us understand how drugs are working. Part of this work has been published (http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1006316). Dr. Stacey Wagner* and Adam Struck* were the lead authors on this study. Future studies on biomarkers of DM will be to understand how changes in the cellular environment affect the biomarkers and understanding the molecular mechanisms that control their behavior.
A better understanding of the proteins that are sequestered by the toxic RNA has lead to a new project in the lab, which is the development of synthetic proteins that could be novel therapeutics for DM. This work is being lead by Promise to Kate (PTK) graduate fellow Melissa Hale* who first began work on this project at the University of Oregon in 2013. Melissa was willing to move 3,000 miles to continue to work on this project at the University of Florida. The PTK fellowship is providing important funding that supports Melissa while we get the first publication out on this exciting work. These results have opened up a new avenue of research for the lab and we recently applied for funding from the National Institutes of Health to support and expand our efforts to engineer novel proteins as potential therapeutics for DM and as tools to understand myotonic dystrophy and more broadly biological processes. The funding from PTK and other foundations is frequently used to obtain preliminary data and publish proof of principal studies that make us competitive in our efforts to obtain funding from national foundations (Muscular Dystrophy Association) and the National Institute of Health.
My colleagues whose research contributions are described above and their current positions
*Dr. Jeremy Logue is currently an Assistant Professor at the Albany Medical College.
*Dr. Blaine Mooers is currently an Assistant Professor at the University of Oklahoma Medical School.
*Dr. Bryan Warf is a Senior Scientist at Myriad Genetics Laboratories.
*Dr. Catherine Matthys is a Physician in Los Angeles CA and affiliated with UCLA Medical Center.
*Dr. Stacey Wagner is the Director of the Applied Bioinformatics and Genomics Master’s Program at the University of Oregon
*Adam Struck is a computational scientist at Oregon Health Sciences University
*Melissa Hale is a PhD student at the University of Florida and PTK Fellow.